- Use udfs only when necessary
- Debug udfs by raising exceptions
- Do not use DataFrame objects inside udfs
- Do not import / define udfs before creating SparkContext
- Defining udfs in a class
User defined function (udf) is a feature in (Py)Spark that allows user to define customized functions with column arguments. This post summarizes some pitfalls when using udfs.
Example of udf:
# dataframe of channelids
import pandas as pd
import numpy as np
import random
from pyspark.sql.functions import udf, lit
from pyspark.sql.types import DoubleType, BooleanType, StringType
import logging
import sys
from pyspark.sql import SQLContext, SparkSession
from pyspark import SparkContext, SQLContext, SparkConf
import os
def square(x):
return float(x*x)
# register udf
square_udf = udf(square, DoubleType())
# sample dataframe
n = 100
numbers = np.random.normal(loc=5, scale=1, size=n)
df_pd = pd.DataFrame({'number': numbers})
df = spark.createDataFrame(df_pd)
# udf usage
df1 = df.withColumn('squared', square_udf(df['number']))
>>> df.show(10)
+------------------+
| number|
+------------------+
| 5.685395575385293|
| 5.202460175305168|
| 5.631595697611453|
|3.5112203388890113|
|3.8605235152919115|
| 4.465308911421083|
|5.3378137934754335|
| 4.137082993915284|
|3.2010281920995576|
| 6.515478687298831|
+------------------+
only showing top 10 rows
>>> df1.show(10)
+------------------+------------------+
| number| squared|
+------------------+------------------+
| 5.685395575385293|32.323722848610664|
| 5.202460175305168| 27.06559187563628|
| 5.631595697611453|31.714870101355828|
|3.5112203388890113|12.328668268227863|
|3.8605235152919115|14.903641812121817|
| 4.465308911421083|19.938983674416537|
|5.3378137934754335|28.492256093816597|
| 4.137082993915284|17.115455698543048|
|3.2010281920995576|10.246581486616162|
| 6.515478687298831| 42.4514625246453|
+------------------+------------------+
only showing top 10 rows
I encountered the following pitfalls when using udfs.
Use udfs only when necessary
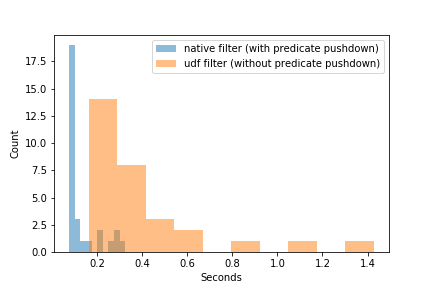
Spark optimizes native operations. One such optimization is predicate pushdown.
A predicate is a statement that is either true or false, e.g., df.amount > 0. Conditions in .where() and .filter() are predicates. Predicate pushdown refers to the behavior that if the native .where() or .filter() are used after loading a dataframe, Spark “pushes” these operations down to the data source level to minimize the amount of data loaded. That is, it will filter then load instead of load then filter.
For udfs, no such optimization exists, as Spark will not and cannot optimize udfs.
Consider a dataframe of orders, individual items in the orders, the number, price, and weight of each item.
# example data
n = 10**5
orderids = sum([[i] * 4 for i in range(n//4)], [])
itemids = list(range(4)) * (n//4)
numbers = [0]*10 + [int(x) for x in np.random.normal(loc=5, scale=1, size=n-10)]
prices = [round(x, 2) for x in np.random.normal(loc=100, scale=20, size=n)]
weights = [round(x, 2) for x in np.random.normal(loc=5, scale=1, size=n)]
df_pd = pd.DataFrame(
{'orderid': orderids,
'itemid': itemids,
'number': numbers,
'price': prices,
'weight': weights}
)
df = spark.createDataFrame(df_pd).select('orderid', 'itemid', 'number', 'price', 'weight')
df.write.parquet('/user/xiaolinfan/demo/df.parquet', mode='overwrite')
>>> df.show(10)
19/10/29 22:30:46 WARN TaskSetManager: Stage 6 contains a task of very large size (381 KB). The maximum recommended task size is 100 KB.
+-------+------+------+------+------+
|orderid|itemid|number| price|weight|
+-------+------+------+------+------+
| 0| 0| 0|105.43| 4.54|
| 0| 1| 0|131.41| 4.88|
| 0| 2| 0|114.43| 6.81|
| 0| 3| 0|141.96| 4.81|
| 1| 0| 0| 94.54| 4.28|
| 1| 1| 0| 93.35| 3.71|
| 1| 2| 0| 82.85| 6.29|
| 1| 3| 0|118.56| 5.15|
| 2| 0| 0|110.24| 4.55|
| 2| 1| 0|103.58| 5.58|
+-------+------+------+------+------+
only showing top 10 rows
Consider reading in the dataframe and selecting only those rows with df.number > 0. Observe the predicate pushdown optimization in the physical plan, as shown by PushedFilters: [IsNotNull(number), GreaterThan(number,0)].
>>> df = spark.read.parquet('/user/xiaolinfan/demo/df.parquet')
>>> df1 = df.filter(df.number > 0)
>>> df1.explain()
== Physical Plan ==
*(2) HashAggregate(keys=[number#72L, weight#74, price#73, itemid#71L, orderid#70L], functions=[])
+- Exchange hashpartitioning(number#72L, weight#74, price#73, itemid#71L, orderid#70L, 200)
+- *(1) HashAggregate(keys=[number#72L, weight#74, price#73, itemid#71L, orderid#70L], functions=[])
+- *(1) Project [orderid#70L, itemid#71L, number#72L, price#73, weight#74]
+- *(1) Filter (isnotnull(number#72L) && (number#72L > 0))
+- *(1) FileScan parquet [orderid#70L,itemid#71L,number#72L,price#73,weight#74] Batched: true, Format: Parquet, Location: InMemoryFileIndex[hdfs://localhost:9000/user/xiaolinfan/demo/df.parquet], PartitionFilters: [], PushedFilters: [IsNotNull(number), GreaterThan(number,0)], ReadSchema: struct<orderid:bigint,itemid:bigint,number:bigint,price:double,weight:double>
Now, instead of df.number > 0, use a filter_udf as the predicate. Observe that there is no longer predicate pushdown in the physical plan, as shown by PushedFilters: [].
>>> filter_udf = udf(lambda x: x > 0, BooleanType())
>>> df2 = df.filter(filter_udf(df['number']))
>>> df2.explain()
== Physical Plan ==
*(3) HashAggregate(keys=[number#72L, weight#74, price#73, itemid#71L, orderid#70L], functions=[])
+- Exchange hashpartitioning(number#72L, weight#74, price#73, itemid#71L, orderid#70L, 200)
+- *(2) HashAggregate(keys=[number#72L, weight#74, price#73, itemid#71L, orderid#70L], functions=[])
+- *(2) Project [orderid#70L, itemid#71L, number#72L, price#73, weight#74]
+- *(2) Filter pythonUDF0#81: boolean
+- BatchEvalPython [<lambda>(number#72L)], [orderid#70L, itemid#71L, number#72L, price#73, weight#74, pythonUDF0#81]
+- *(1) FileScan parquet [orderid#70L,itemid#71L,number#72L,price#73,weight#74] Batched: true, Format: Parquet, Location: InMemoryFileIndex[hdfs://localhost:9000/user/xiaolinfan/demo/df.parquet], PartitionFilters: [], PushedFilters: [], ReadSchema: struct<orderid:bigint,itemid:bigint,number:bigint,price:double,weight:double>
Debug udfs by raising exceptions
Programs are usually debugged by raising exceptions, inserting breakpoints (e.g., using debugger), or quick printing/logging.
Debugging (Py)Spark udfs requires some special handling.
Consider the same sample dataframe created before. Suppose we want to calculate the total price and weight of each item in the orders via the udfs get_item_price_udf() and get_item_weight_udf(). Suppose further that we want to print the number and price of the item if the total item price is no greater than 0. (We use printing instead of logging as an example because logging from Pyspark requires further configurations, see here).
# example data
df = spark.read.parquet('/user/xiaolinfan/demo/df.parquet')
# debugging
def get_item_price(number, price):
item_price = float(number * price)
if item_price <= 0.:
msg = 'number = {}, price = {}'.format(number, price)
print(msg)
return item_price
def get_item_weight(number, weight):
item_weight = float(number * weight)
return item_weight
get_item_price_udf = udf(get_item_price, DoubleType())
get_item_weight_udf = udf(get_item_weight, DoubleType())
df_item = df.withColumn('item_price', get_item_price_udf(df['number'], df['price']))\
.withColumn('item_weight', get_item_price_udf(df['number'], df['weight']))
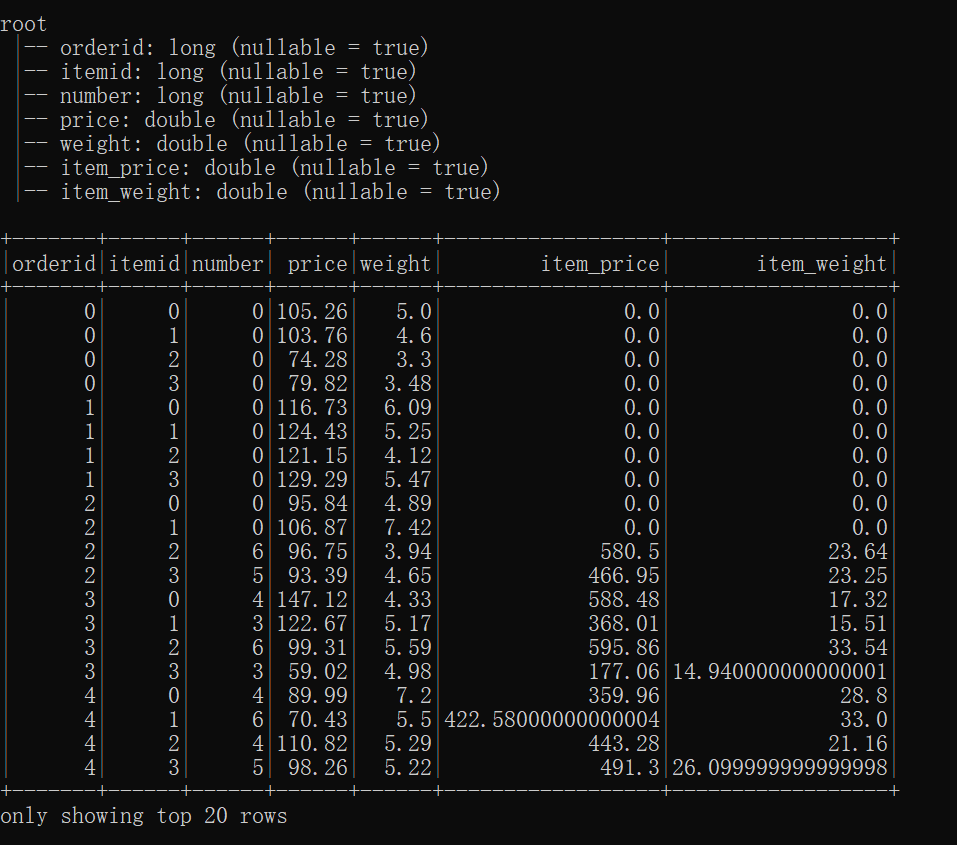
df_item.printSchema()
df_item.show()
Submitting this script via spark-submit --master yarn generates the following output. Observe that the the first 10 rows of the dataframe have item_price == 0.0, and the .show() command computes the first 20 rows of the dataframe, so we expect the print() statements in get_item_price_udf() to be executed. However, they are not printed to the console.
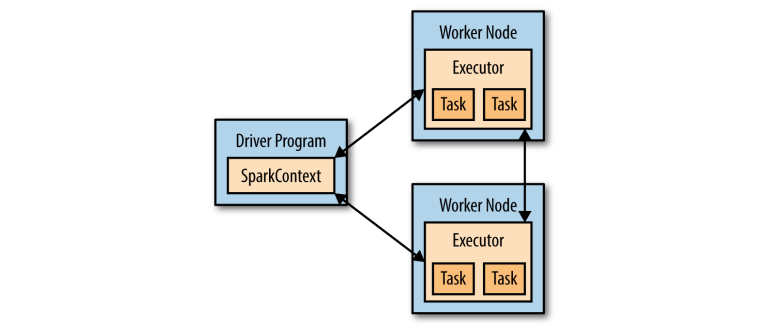
This can be explained by the nature of distributed execution in Spark (see here). In short, objects are defined in driver program but are executed at worker nodes (or executors). In particular, udfs are executed at executors. Thus, in order to see the print() statements inside udfs, we need to view the executor logs.
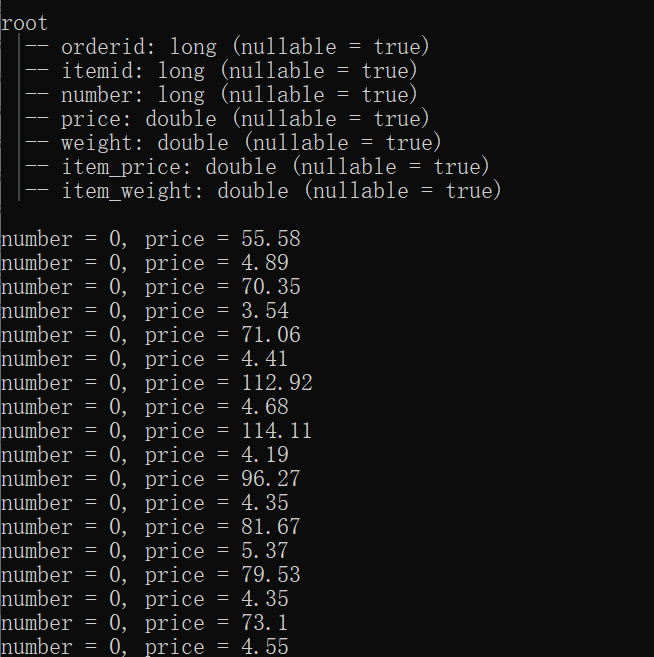
Another way to validate this is to observe that if we submit the spark job in standalone mode without distributed execution, we can directly see the udf print() statements in the console:
There are a few workarounds.
Yarn commands
- Enable yarn log aggregation by setting
<property>
<name>yarn.log-aggregation-enable</name>
<value>true</value>
</property>
in yarn-site.xml in $HADOOP_HOME/etc/hadoop/.
- View executor logs via
yarn logs -applicationId <application_id>, as instructed here.application_idcan be found in the resource manager UI
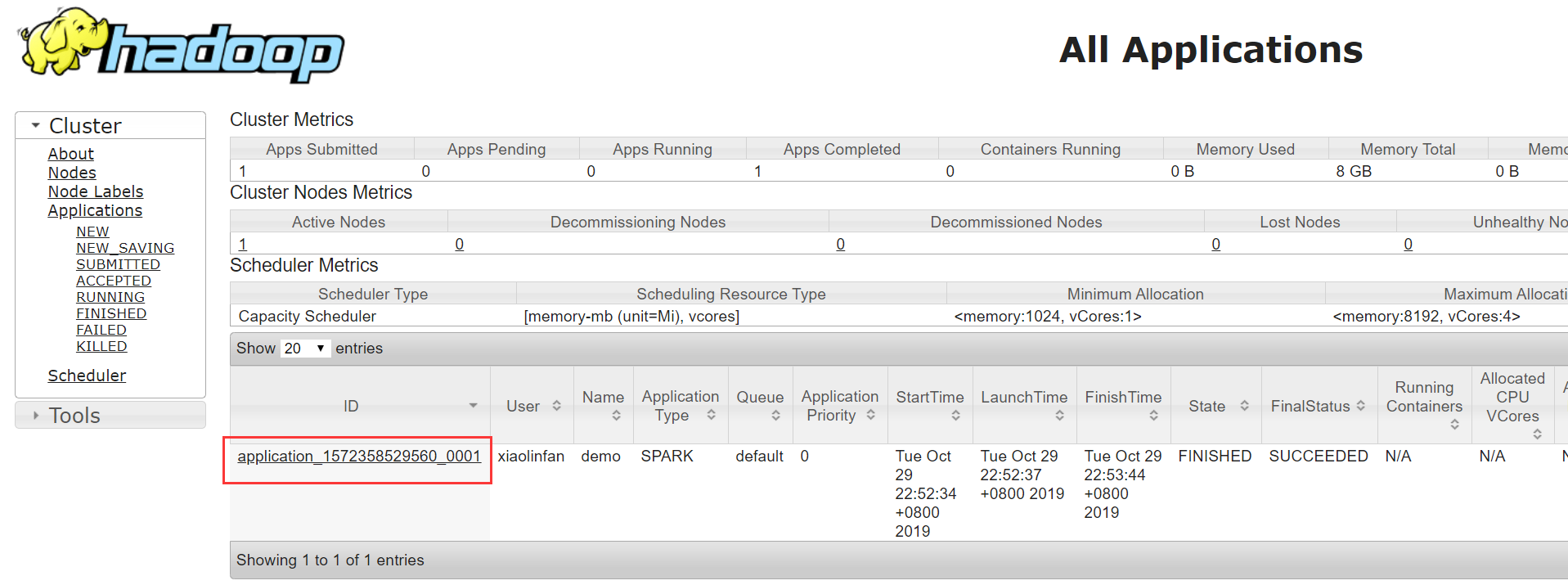
or via the command yarn application -list -appStates ALL (-appStates ALL shows applications that are finished).
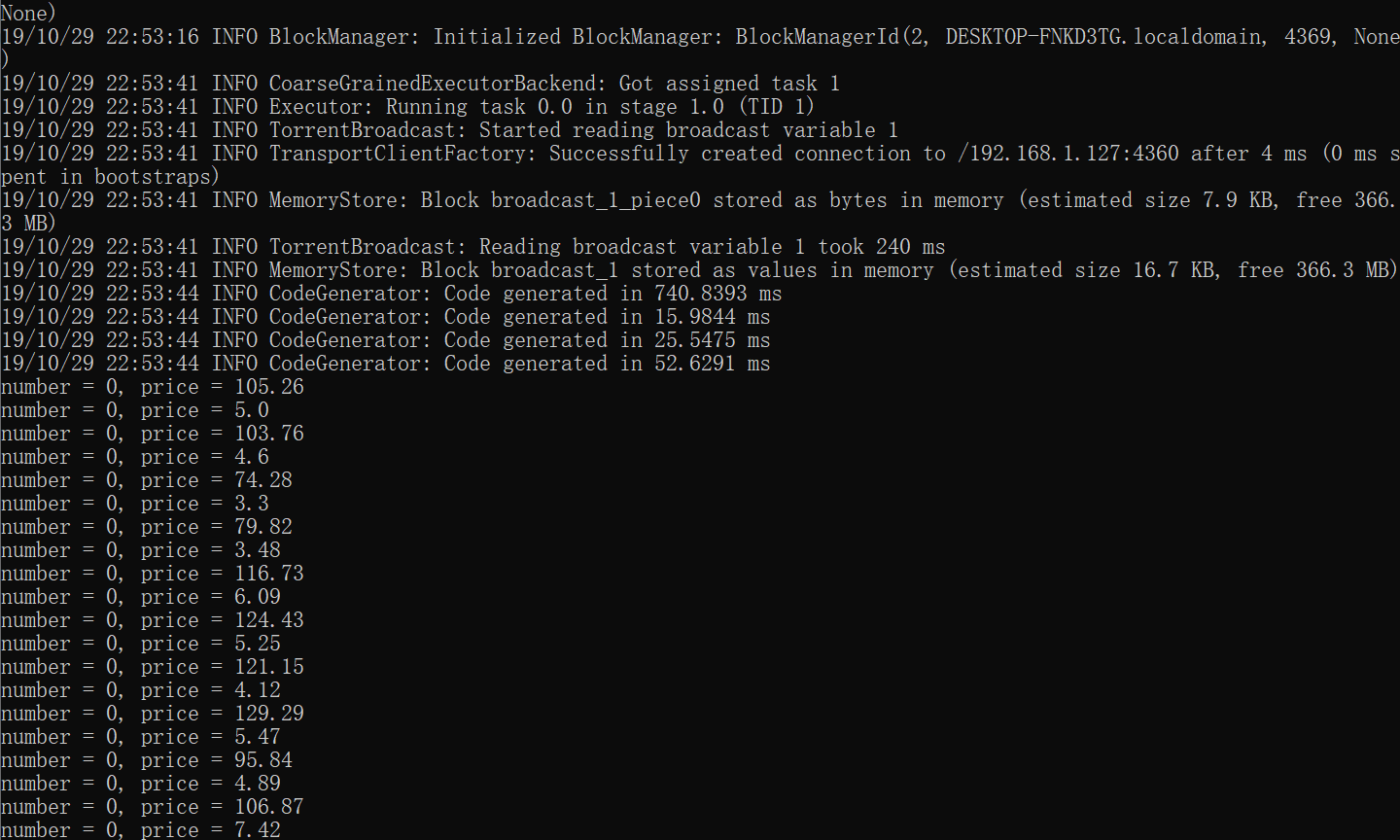
Note: To see that the above is the log of an executor and not the driver, can view the driver ip address at yarn application -status <application_id>. Usually, the container ending with 000001 is where the driver is run.
This method is straightforward, but requires access to yarn configurations. This could be not as straightforward if the production environment is not managed by the user.
Raise exceptions
Another way to show information from udf is to raise exceptions, e.g.,
def get_item_price(number, price):
item_price = float(number * price)
if item_price <= 0.:
msg = 'number = {}, price = {}'.format(number, price)
raise Exception(msg)
return item_price
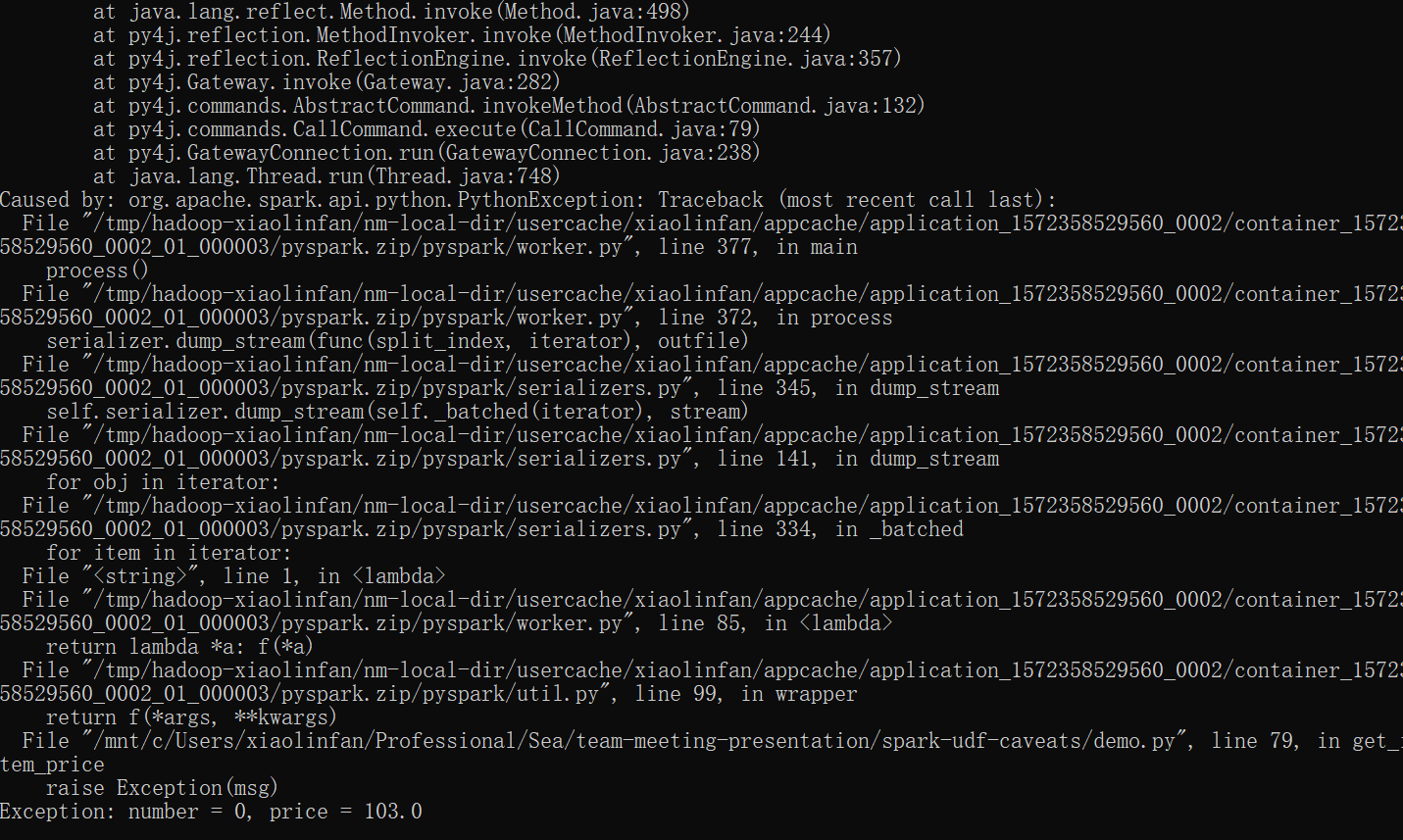
This method is independent from production environment configurations. But the program does not continue after raising exception.
Return message with output
Yet another workaround is to wrap the message with the output, as suggested here, and then extract the real output afterwards.
Do not use DataFrame objects inside udfs
Serialization is the process of turning an object into a format that can be stored/transmitted (e.g., byte stream) and reconstructed later.
E.g., serializing and deserializing trees:
Because Spark uses distributed execution, objects defined in driver need to be sent to workers. This requires them to be serializable. In particular, udfs need to be serializable.
Consider a dataframe of orderids and channelids associated with the dataframe constructed previously. Suppose we want to add a column of channelids to the original dataframe. We do this via a udf get_channelid_udf() that returns a channelid given an orderid (this could be done with a join, but for the sake of giving an example, we use the udf).
# dataframe of channelids
n = 100
orderids = list(set(sum([[i] * 4 for i in range(n//4)], [])))
channelids = np.random.choice(list(range(5)),len(orderids))
channel_df_pd = pd.DataFrame(
{'orderid': orderids,
'channelid': channelids}
)
channel_df = spark.createDataFrame(channel_df_pd).select('orderid', 'channelid')
channel_df.printSchema()
channel_df.show()
channel_df.registerTempTable('channel_df')
# get channelid from orderid
def get_channelid(orderid):
channelid = channel_df.where(channel_df.orderid == orderid).select('channelid').collect()[0]['channelid']
return str(channelid)
# register as udf
get_channelid_udf = udf(get_channelid, StringType())
# apply udf
df_channelid = df.withColumn('channelid', get_channelid_udf(df['orderid']))
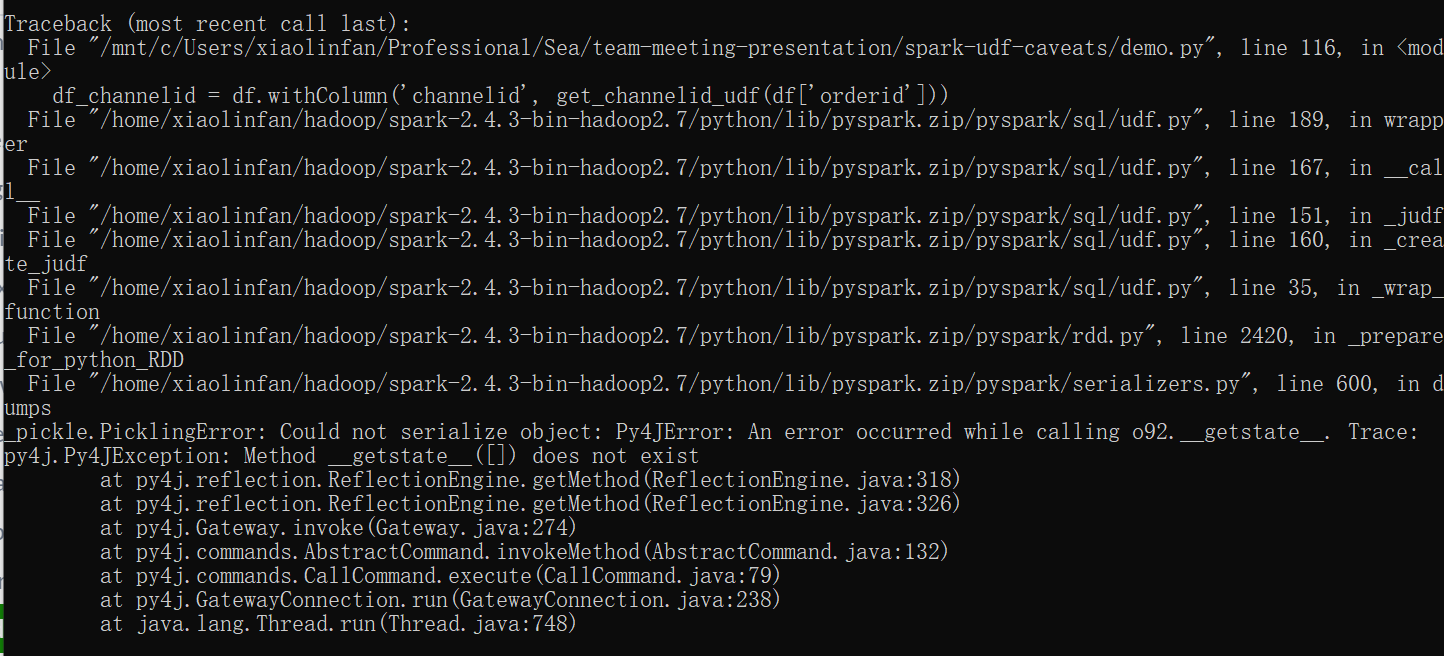
This is because the Spark context is not serializable. (Though it may be in the future, see here.) Since udfs need to be serialized to be sent to the executors, a Spark context (e.g., dataframe, querying) inside an udf would raise the above error.
Some workarounds include:
- If the query is simple, use join.
- If the query is too complex to use join and the dataframe is small enough to fit in memory, consider converting the Spark dataframe to Pandas dataframe via
.toPandas()and perform query on the pandas dataframe object. - If the object concerned is not a Spark context, consider implementing Java’s Serializable interface (e.g., in Scala, this would be
extend Serializable).
Do not import / define udfs before creating SparkContext
Spark udfs require SparkContext to work. So udfs must be defined or imported after having initialized a SparkContext. Otherwise, the Spark job will freeze, see here.
Defining udfs in a class
If udfs are defined at top-level, they can be imported without errors.
If udfs need to be put in a class, they should be defined as attributes built from static methods of the class, e.g.,
class Function():
def __init__(self):
self.foo_udf = F.udf(Function.foo, T.DoubleType())
@staticmethod
def foo():
return .5
otherwise they may cause serialization errors.
An explanation is that only objects defined at top-level are serializable. These include udfs defined at top-level, attributes of a class defined at top-level, but not methods of that class (see here).